Getting Started
To install the nybem package, install from GitHub using
the remotes package:
remotes::install_github(repo = "MVR-GIS/nybem")Load Required Packages
##
## Attaching package: 'nybem'## The following object is masked from 'package:base':
##
## ifelse## Warning: package 'raster' was built under R version 4.2.1## Loading required package: sp## Warning: package 'sp' was built under R version 4.2.1List the NYBEM Models
The nybem package contains the models created for the
ecological modeling section of the NTBEM feasibility study. Notice that
the nybem::nybem_submodels list contains a pair of data
frames for each submodel in the study. Each pair of dataframes is named
for the submodel it represents (i.e., NYBEM.fresh.tid =
Freshwater Tidal Zone habitat model). The first data frame contains the
habitat variables and their matching suitability index values, while the
second data frame (*_labels) contains the labels used in
plotting.
# Load the model data
nybem_submodels <- nybem::nybem_submodels
# View the available submodels
names(nybem_submodels)## [1] "NYBEM.fresh.tid" "NYBEM.fresh.tid_labels"
## [3] "NYBEM.est.int" "NYBEM.est.int_labels"
## [5] "NYBEM.est.sub.hard" "NYBEM.est.sub.hard_labels"
## [7] "NYBEM.est.sub.soft.sav" "NYBEM.est.sub.soft.sav_labels"
## [9] "NYBEM.est.sub.soft.clam" "NYBEM.est.sub.soft.clam_labels"
## [11] "NYBEM.mar.int" "NYBEM.mar.int_labels"
## [13] "NYBEM.mar.sub" "NYBEM.mar.sub_labels"
## [15] "NYBEM.mar.deep" "NYBEM.mar.deep_labels"View the Suitability Index data frame
# View the Freshwater Tidal model
# View table of habitat variables and their suitability index values
nybem_submodels$NYBEM.fresh.tid## salinity.per salinity.SIV veg.cover.per veg.cover.SIV deposition
## 1 0 1 0 0.1 1
## 2 20 0 57 0.5 2
## 3 100 0 100 1.0 5
## deposition.SIV
## 1 0
## 2 1
## 3 1View the Suitability Index model labels
nybem_submodels$NYBEM.fresh.tid_labels## model label variable
## 1 Freshwater, Tidal Salinity duration (% time > 0.5 psu) salinity
## 2 Freshwater, Tidal Vegetation Cover (%) veg.cover
## 3 Freshwater, Tidal Relative Depth depositionPlot the Suitability Index Values
# Freshwater, Tidal
nybem::HSIplotter(nybem_submodels$NYBEM.fresh.tid,
xlab = nybem_submodels$NYBEM.fresh.tid_labels$label,
ylab = nybem_submodels$NYBEM.fresh.tid_labels$variable)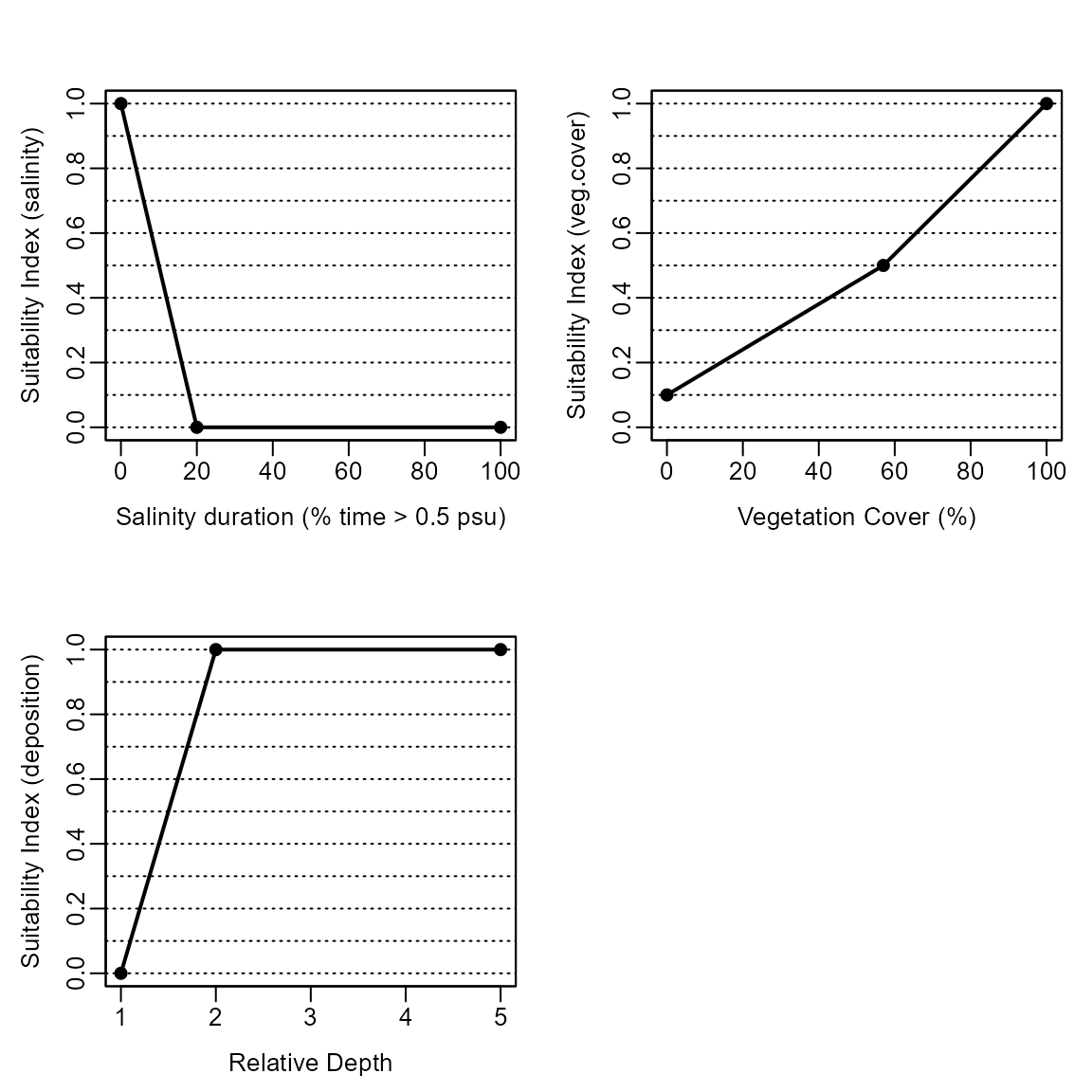
Create Site Observations
# Single numeric site observations
salinity <- 40
veg.cover <- 100
deposition <- 0
obs_1 <- list(salinity, veg.cover, deposition)
obs_1## [[1]]
## [1] 40
##
## [[2]]
## [1] 100
##
## [[3]]
## [1] 0
# Raster site observations
salinity_ras <- raster::raster(ncol = 2, nrow = 2, vals = rep_len(40, 4))
veg.cover_ras <- raster::raster(ncol = 2, nrow = 2, vals = rep_len(100, 4))
deposition_ras <- raster::raster(ncol = 2, nrow = 2, vals = rep_len(0, 4))
obs_2 <- list(salinity_ras, veg.cover_ras, deposition_ras)
obs_2## [[1]]
## class : RasterLayer
## dimensions : 2, 2, 4 (nrow, ncol, ncell)
## resolution : 180, 90 (x, y)
## extent : -180, 180, -90, 90 (xmin, xmax, ymin, ymax)
## crs : +proj=longlat +datum=WGS84 +no_defs
## source : memory
## names : layer
## values : 40, 40 (min, max)
##
##
## [[2]]
## class : RasterLayer
## dimensions : 2, 2, 4 (nrow, ncol, ncell)
## resolution : 180, 90 (x, y)
## extent : -180, 180, -90, 90 (xmin, xmax, ymin, ymax)
## crs : +proj=longlat +datum=WGS84 +no_defs
## source : memory
## names : layer
## values : 100, 100 (min, max)
##
##
## [[3]]
## class : RasterLayer
## dimensions : 2, 2, 4 (nrow, ncol, ncell)
## resolution : 180, 90 (x, y)
## extent : -180, 180, -90, 90 (xmin, xmax, ymin, ymax)
## crs : +proj=longlat +datum=WGS84 +no_defs
## source : memory
## names : layer
## values : 0, 0 (min, max)Calculate Suitability Index Values
# Suitability Index value for single numeric site observations
siv_fresh.tid_obs_1 <- nybem::SIcalc(SI = nybem_submodels$NYBEM.fresh.tid,
input_proj = obs_1)
siv_fresh.tid_obs_1## [[1]]
## [1] 0
##
## [[2]]
## [1] 1
##
## [[3]]
## [1] 0
# Suitability Index value for raster site observations
siv_fresh.tid_obs_2 <- nybem::SIcalc(SI = nybem_submodels$NYBEM.fresh.tid,
input_proj = obs_2)
siv_fresh.tid_obs_2## [[1]]
## class : RasterLayer
## dimensions : 2, 2, 4 (nrow, ncol, ncell)
## resolution : 180, 90 (x, y)
## extent : -180, 180, -90, 90 (xmin, xmax, ymin, ymax)
## crs : +proj=longlat +datum=WGS84 +no_defs
## source : memory
## names : layer
## values : 0, 0 (min, max)
##
##
## [[2]]
## class : RasterLayer
## dimensions : 2, 2, 4 (nrow, ncol, ncell)
## resolution : 180, 90 (x, y)
## extent : -180, 180, -90, 90 (xmin, xmax, ymin, ymax)
## crs : +proj=longlat +datum=WGS84 +no_defs
## source : memory
## names : layer
## values : 1, 1 (min, max)
##
##
## [[3]]
## class : RasterLayer
## dimensions : 2, 2, 4 (nrow, ncol, ncell)
## resolution : 180, 90 (x, y)
## extent : -180, 180, -90, 90 (xmin, xmax, ymin, ymax)
## crs : +proj=longlat +datum=WGS84 +no_defs
## source : memory
## names : layer
## values : 0, 0 (min, max)Calculate Habitat Suitability Index
# Calculate HSI for single numeric site observations
hsi_fresh.tid_obs_1 <- nybem::HSIcalc(si_list = siv_fresh.tid_obs_1)
hsi_fresh.tid_obs_1## [1] 0.3333333
# Calculate HSI for raster site observations
hsi_fresh.tid_obs_2 <- nybem::HSIcalc(si_list = siv_fresh.tid_obs_2)
hsi_fresh.tid_obs_2## class : RasterLayer
## dimensions : 2, 2, 4 (nrow, ncol, ncell)
## resolution : 180, 90 (x, y)
## extent : -180, 180, -90, 90 (xmin, xmax, ymin, ymax)
## crs : +proj=longlat +datum=WGS84 +no_defs
## source : memory
## names : layer
## values : 0.3333333, 0.3333333 (min, max)